About Metadata
Already understand what metadata is, but trying to figure out how to find it in the portal? See How to find and download metadata files.
When data is uploaded into the AD Knowledge Portal, it must be done so in a way that makes it easily accessible and readable to the users who download it. This is where metadata comes in.
Metadata is standardized information provided along with data that helps with data organization and description. It is, essentially, data about the data.
Metadata is useful for you and everyone else that has a need for discovering, accessing, and/or using the corresponding data or repository the data is stored in. Therefore, effort is placed on the importance of metadata being understandable to a diverse set of users. This gives you the ability to interpret the data in a well-rounded, comprehensive way that tells the whole picture of the data.
What is the purpose of metadata?
Metadata serves many purposes:
Data understandability
Metadata gives context to the data, allowing it to be understood by others, including those who were not involved in the data generation process. For example, descriptive metadata such as the study name, the assay performed, tissue type, species, etc., provides general information about the data that allows a user, such as a bioinformatician, to decide if they can reuse the data for analysis or other purposes.
Data discovery and accessibility
Because metadata is provided in a standardized format, it allows the data to be searchable by various metadata elements, as well as accessible, all in an organized way. This standardization can greatly improve the accuracy of a search and reduce ambiguity.
Data interoperability
The standardized nature of metadata allows the data to be efficiently integrated with other applications.
Data reuse
All of the aforementioned purposes of metadata contribute to a data’s ability to be re-used. If data is not understandable, discoverable, accessible, or interoperable, it will be difficult or even impossible to reuse.
What is the structure of metadata?
All data and metadata that gets uploaded into Synapse is curated by the AD-DCC to ensure the metadata properly allows for data usability. In most cases, we require three things for every set of data:
The data itself, of course
A description of the study and methods used to generate the data
The metadata
While a subset of metadata is stored as annotations on the data files themselves, the complete metadata are stored within a separate set of files (in the form of .CSV, or comma-separated values—in a program such as Microsoft Excel). These metadata include extra details and about individuals, specimens (or biosamples) from the individuals, and the assay performed on the specimens.
We can think of metadata as being four tables, which get submitted to the portal as four files:

![]() As a data user, you only need to know about the first three files. The manifest file is handled by the data uploader and is not applicable to you as a data user. If you’re a data contributor, find more information on creating and uploading the manifest (and all other metadata) here.
As a data user, you only need to know about the first three files. The manifest file is handled by the data uploader and is not applicable to you as a data user. If you’re a data contributor, find more information on creating and uploading the manifest (and all other metadata) here.
The manifest file includes metadata about the files themselves. The data uploader uses this file to provide a set of metadata variables for the purpose of file annotations. These annotations are what allow you to perform queries while exploring data, in order to find and download the specific data you need.
What is contained in each metadata file?
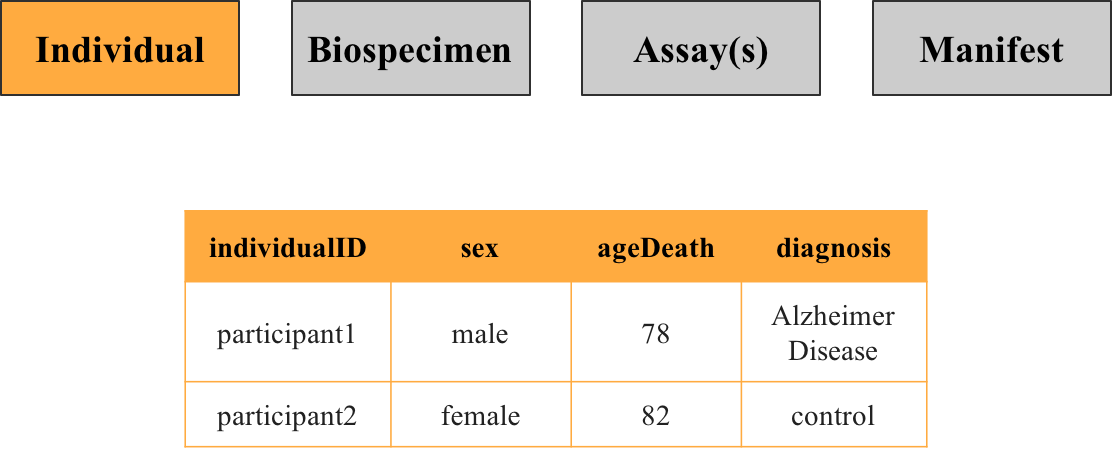
1. Individual metadata
This file contains information about the participants in the study, such as their sex and diagnosis. Each row corresponds to a unique individual identifiable by the key individualID.
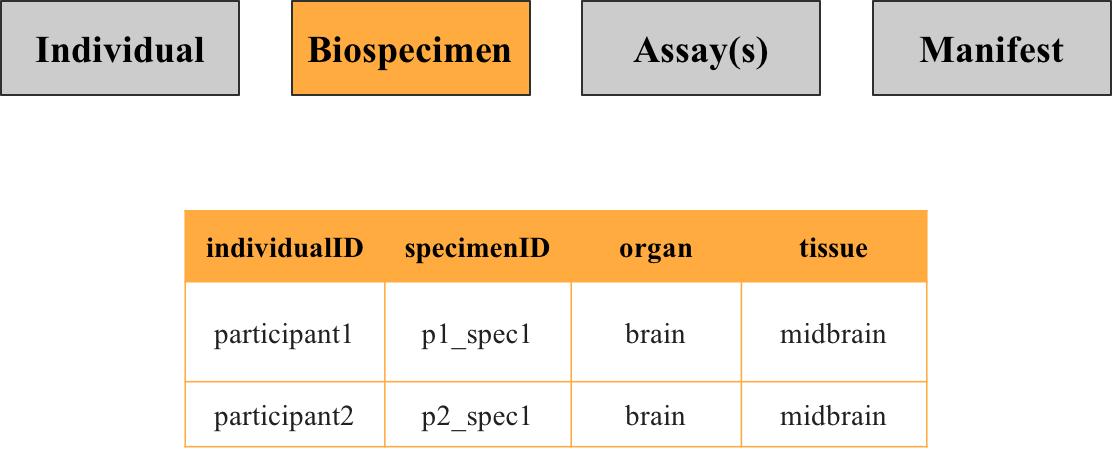
2. Biospecimen metadata
This file contains information about the specimens that were collected from participant in the study, such as what organ and tissue the specimen was taken from. While each row corresponds to a unique specimen identified by the key specimenID, there may be multiple specimens per individual.
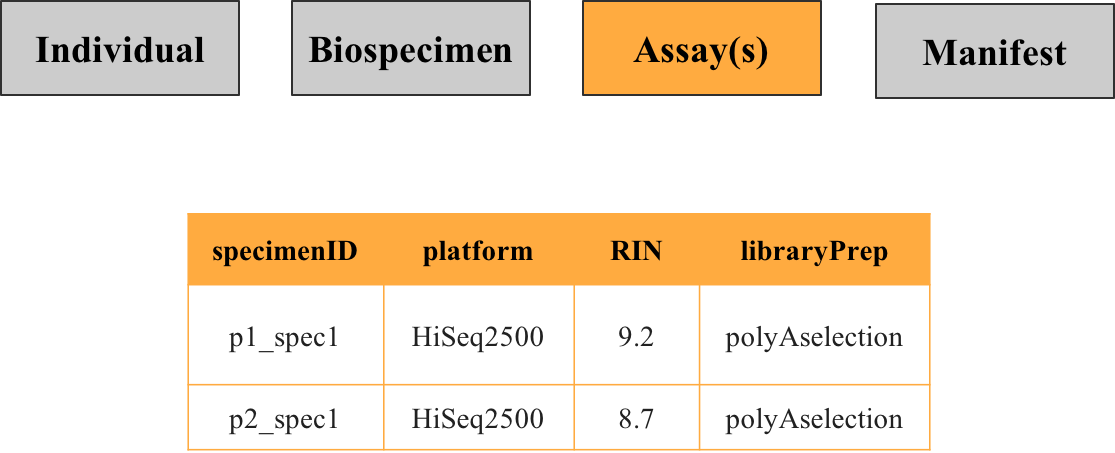
3. Assay(s) metadata
This file contains information about what was done to the specimen in order to generate the data, such as the technology used, batch [processing, quality control metrics, etc.. For example, for RNA sequencing the metadata would specify what platform was used and how the specimen was prepared. If specimens or individuals were characterized across several assay types, there may be more than one assay metadata file.
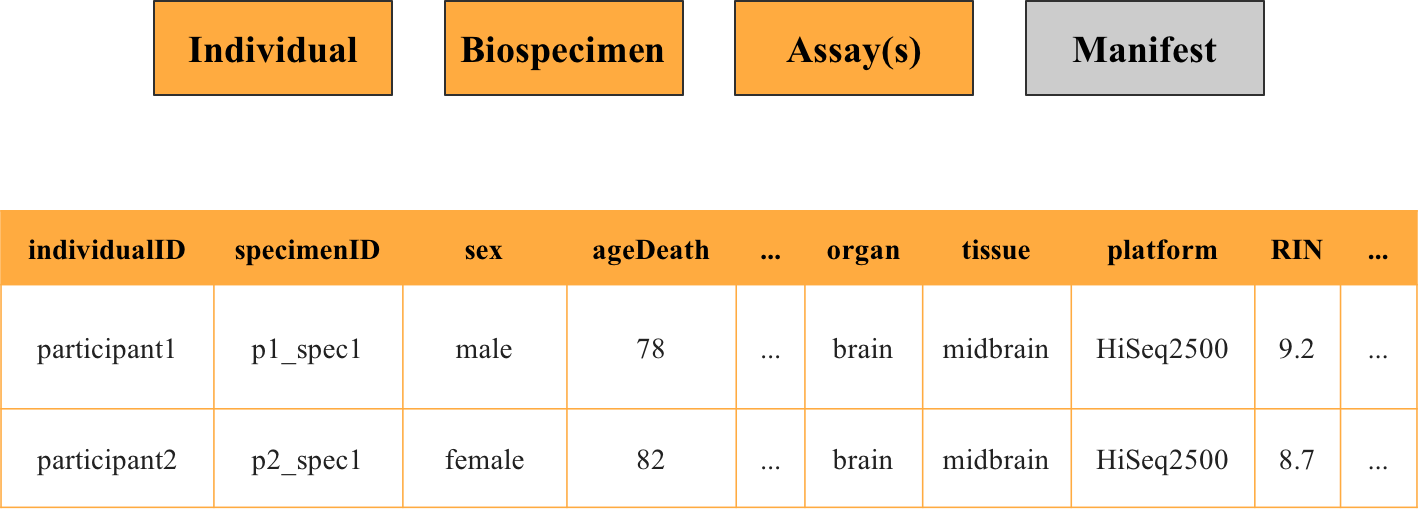
Can you join these metadata files together?
Yes, you can! Metadata files of the individual, biospecimen, and assay type can be joined together on the keys individualID and specimenID, and can be combined with the data itself, to get a complete picture with details on each file (such as seen in the example below)
For helpful information and instructions on how to actually find and identify metadata within the portal, see How to find and download metadata files.
Further, you can see an example of this in action by reading our Use Case #3: Accessing and Joining Metadata Files With Data
Metadata dictionary
The metadata dictionary provides the proper terms that you need to know in order to successfully query for data as a user, or properly contribute data as a contributor. As a user, knowing this information will allow you to search for and find the data you need.
If you’re a data contributor, you can find more information on annotations and how to assign them to your uploaded data here, and information on creating metadata here.
In general, the metadata dictionary is useful if you are:
A user who needs to know what terms in the annotations and metadata files mean so you can properly search for and find the data you need.
A contributor who needs to know what the accepted values are for a given key. For example, if you used a HiSeq 2000 machine to generate your data, then you would need to know that the accepted value for that machine is HiSeq2000 (no space). You also need to know that it goes in the column with the key platform.
A curator who needs to know what keys/values are currently available in order to direct contributors to use them, or gauge a need for any additional ones.
How to access the metadata dictionary
In order to access the metadata dictionary, you must be logged into Synapse. If you are not already logged into Synapse when you land on this page, you will be prompted to do so. Click the Synapse link provided in the pop-up window to log in to Synapse, and then revisit or refresh the metadata dictionary—you should now be able to access it.
Additionally, if you have an ad blocker installed on your browser, you must disable it for this website. To do so, click the ad blocker icon in the top right corner of your browser, and clock the toggle option to disable ads on this page.
How to use the metadata dictionary
To make use of the metadata dictionary, the various search fields are very helpful. First, notice that there’s a global search bar at the top of the page. You can use this to search for any field. Then, notice that each column has its own dedicated search bar. You can search for specific terms within each column to narrow down the list. You can find a breakdown of each column below. A final note is that what you see on your screen is just a subset of metadata—just a few rows out of hundreds. Scroll down to the bottom to see a page navigator, where you can jump to a different page/row in the table.
Here’s a breakdown of the various columns in the dictionary:
Key: Name of metadata term.
Key description: Definition of the metadata term.
Type: Expected data type, such as string or integer, for values associated with the metadata term.
Value: For keys that have the type string, this will either be empty if any text is accepted or will be a specific text value if there are a limited set of values to choose from.
Value description: Description of the value, if any.
Source: Link or name of resource used for the value description, if any.
Module: The group of terms the key is associated with.
